Tutorial
We’ll show here how to explain molecular property prediction tasks without access to the gradients or any properties of a molecule. To set-up this activity, we need a black box model. We’ll use something simple here – the model is classifier that says if a molecule as an alcohol (1) or not (0). Let’s implement this model first
from rdkit import Chem
from rdkit.Chem.Draw import IPythonConsole
# set-up rdkit drawing preferences
IPythonConsole.ipython_useSVG = True
IPythonConsole.drawOptions.drawMolsSameScale = False
def model(smiles):
mol = Chem.MolFromSmiles(smiles)
match = mol.GetSubstructMatches(Chem.MolFromSmarts('[O;!H0]'))
return 1 if match else 0
Let’s now try it out on some molecules
smi = 'CCCCCCO'
print('f(s)', model(smi))
Chem.MolFromSmiles(smi)
f(s) 1
smi = 'OCCCCCCO'
print('f(s)', model(smi))
Chem.MolFromSmiles(smi)
f(s) 1
smi = 'c1ccccc1'
print('f(s)', model(smi))
Chem.MolFromSmiles(smi)
f(s) 0
Counterfacutal explanations
Let’s now explain the model - pretending we don’t know how it works - using counterfactuals
import exmol
instance = 'CCCCCCO'
space = exmol.sample_space(instance, model, batched=False)
cfs = exmol.cf_explain(space, 1)
exmol.plot_cf(cfs)

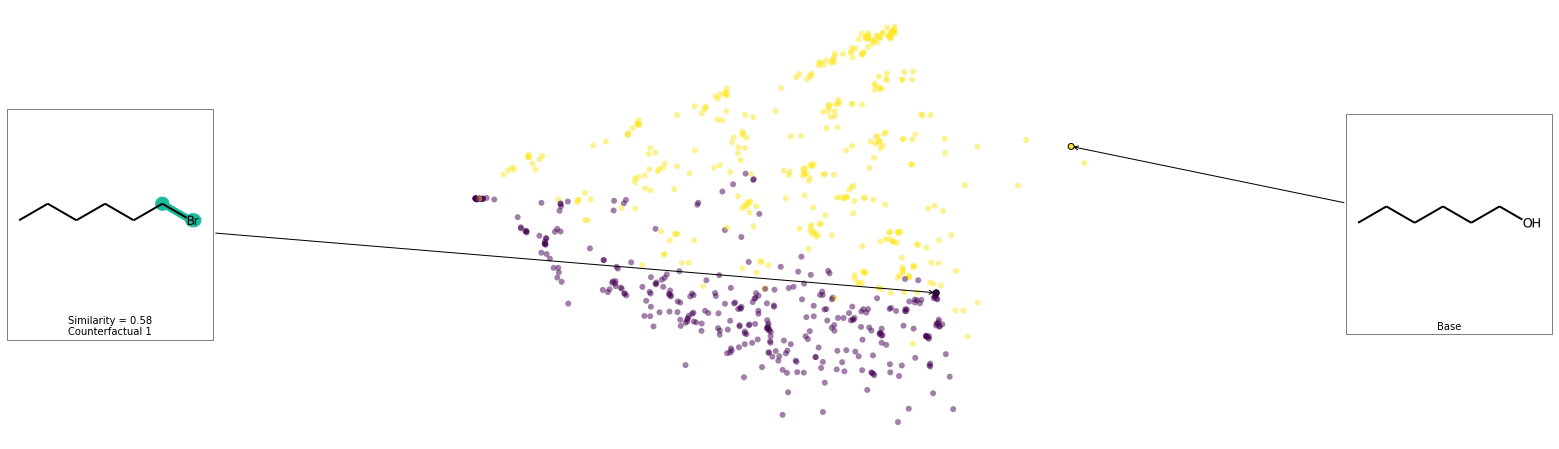
We can see that removing the alcohol is the smallest change to affect the prediction of this molecule. Let’s see the space and look at where these counterfactuals are.
exmol.plot_space(space, cfs)
Explain using substructures
Now we’ll try to explain our model using substructures.
exmol.lime_explain(space)
exmol.plot_descriptors(space)
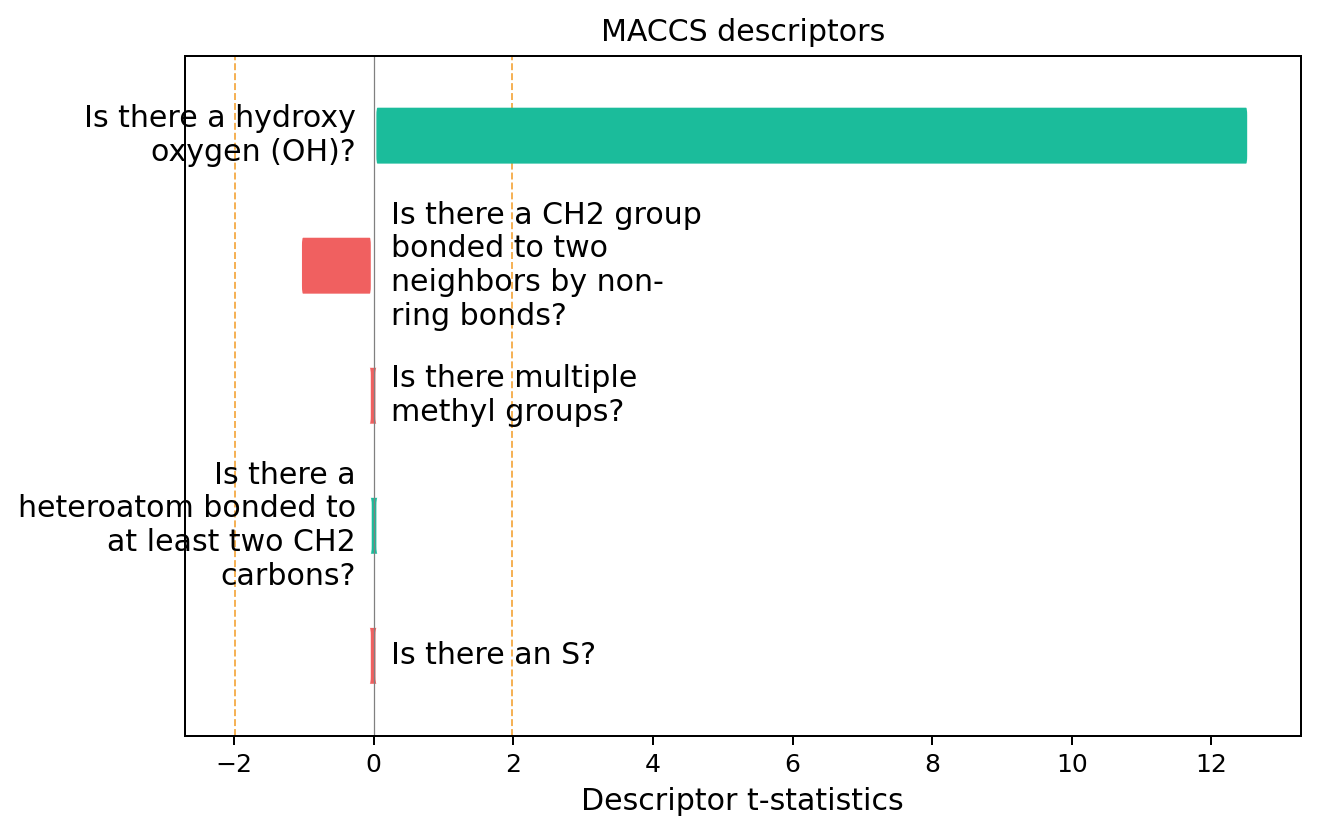
This seems like a pretty clear explanation. Let’s take a look at using substructures that are present in the molecule
import skunk
exmol.lime_explain(space, descriptor_type='ECFP')
svg = exmol.plot_descriptors(space, return_svg=True)
skunk.display(svg)
exmol.plot_utils.similarity_map_using_tstats(space[0])
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Input In [9], in <cell line: 4>()
2 exmol.lime_explain(space, descriptor_type='ECFP')
3 svg = exmol.plot_descriptors(space, return_svg=True)
----> 4 skunk.display(svg)
5 exmol.plot_utils.similarity_map_using_tstats(space[0])
File /opt/hostedtoolcache/Python/3.8.12/x64/lib/python3.8/site-packages/skunk/__init__.py:64, in display(svg)
62 import IPython.display as display
63 import base64
---> 64 data = base64.b64encode(svg.encode('utf8'))
65 display.display(display.HTML(
66 '<img src=data:image/svg+xml;base64,' + data.decode() + '>'))
AttributeError: 'NoneType' object has no attribute 'encode'
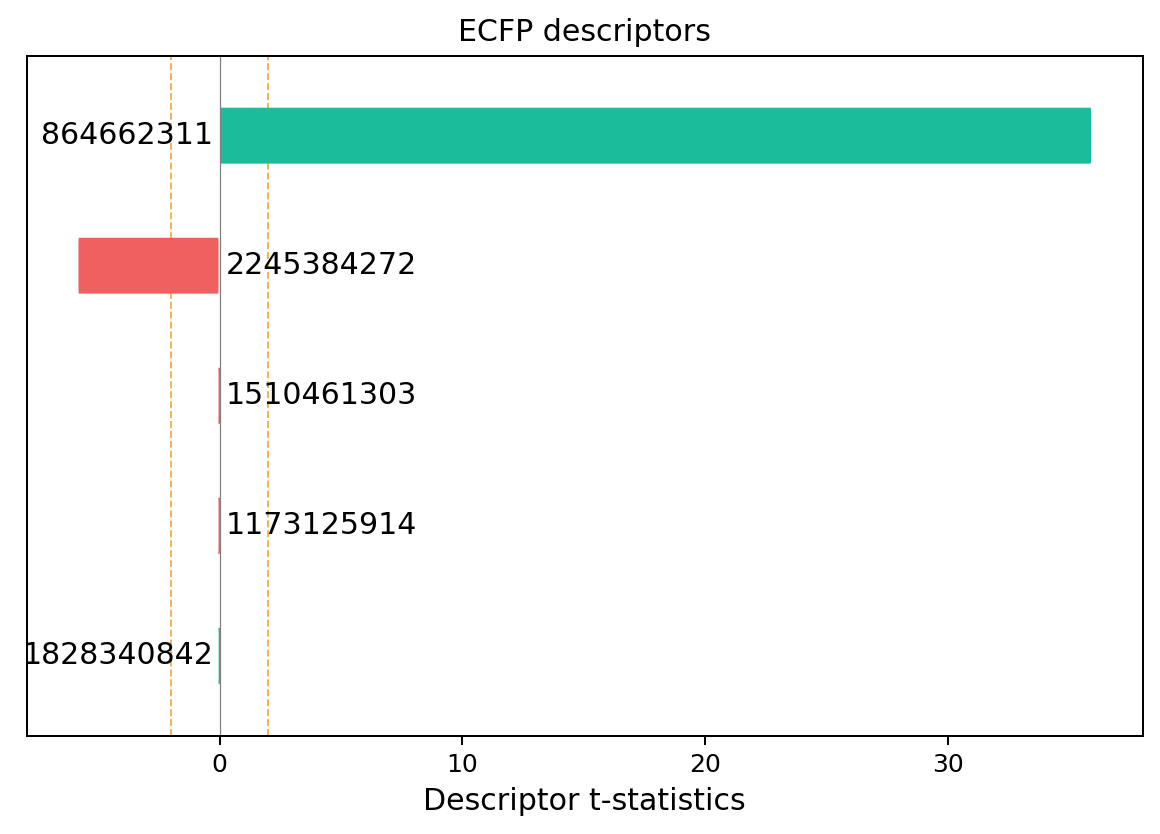
We can see that most of the model is explained from the presence of the alcohol group - as expected